Trusted Research Environments (TRE) Green Paper¶
A strategy to build public trust and meet changing health data science needs
Executive Summary¶
The UK Health Data Research Alliance is an independent alliance of data providers, custodians and curators dedicated to improving human health by maximising the potential of multiple forms of data at scale.
Our vision is that every health and care interaction and research endeavour will be enhanced by access to large scale data and advanced analytics.
1.Why does this matter?
Even before the COVID-19 pandemic, challenges to human health and health system sustainability have been increasing across the world. Whilst heart disease, stroke and cancer still account for nearly two thirds of all deaths globally, increasingly peope living with multiple diseases and long-term conditions. These affect us deeply -- they change the lives of those with the diseases and those who care for them. By making health data available to researchers, we can develop a better understanding of these diseases and find ways to prevent, diagnose, treat and manage or cure them. It is also essential for tackling the direct and indirect impact of pandemics and other public health challenges.
2.Why is the UK the best place to do this?
The UK has some of the richest health data anywhere in the world. With the NHS it is feasible to collect longitudinal health data on a large and diverse population, and to make national-scale improvements to health and care. Combined with unique research expertise, outstanding talent in the NHS and universities, and vibrant life sciences and technology industries, the UK has an unprecedented opportunity to use data at scale to drive innovation, grow the UK industry base and improve the long-term health of the public. For example, the RECOVERY trial1 is currently the world's largest trial of COVID-19 drugs2.
3. How do we ensure this happens in a safe way that retains and enhances public trust?
Research conducted by Understanding Patient Data identified that people are generally comfortable with anonymised data from medical records being used for improving health, care and services, and for research, provided there is a public benefit. 3 The more informed people feel, the more they are likely to support these uses. However, people are more likely to be uncomfortable with the idea of commercial companies accessing their health data, and there are concerns about information being passed on for marketing or insurance purposes. There are recent examples of projects which have been widely reported in the media and may have increased public concern about health data use4.
The UK Health Data Research Alliance (the 'Alliance') is committed to an approach to data access based primarily around Trusted (Trustworthy) Research Environments (TREs); with appropriate robust and independent TRE accreditation, monitoring and auditing.
Adopting this approach would be a way of addressing these public concerns and enhancing public confidence in the use of health data for research in the UK. For researchers, it would involve a significantly different way of working with these wider health datasets (though would not necessarily change the modes of access to existing research cohort data). Rather than extracts of individual level data being 'released', TREs would provide access to a secure analytics environment (i.e. a safe setting) where researchers could bring analysis algorithms to the data.
To achieve this shift, it will be therefore be necessary to mitigate concerns and gain and sustain the trust of:
-
The public and patients through improved explanations of the benefits and risks associated with using health data as well greater transparency and lay descriptions of the technical solutions being implemented.
-
Data custodians who must be willing to make data available for linkage and use in TREs.
-
Researchers, many of whom are used to a data release model, who may be concerned at the detrimental impact on researcher productivity from working in a TRE environment.
There are now multiple examples of TREs operating successfully in this way, both for healthcare data and other potentially sensitive data, and the responses to the COVID-19 pandemic has accelerated this shift5. Examples include, but are not limited to:
-
Scotland Data Safe Haven programme6
-
UK Secure eResearch Platform in Wales7
-
Genomics England Research Environment8
-
UK Data Service Secure Lab9
-
NHS Digital TRE for England10
-
OpenSAFELY11
There are a growing number of practical and cost saving benefits to this approach. It can maximise the utilisation of High-Performance Computing, whilst avoiding the costs of transferring and storing duplicates of increasingly large datasets, particularly imaging and genomic modalities. It also avoids the liabilities for researchers from having to ensure security of downloaded datasets.
The initial consultation on the draft of this paper indicated broad support for the direction of travel, especially from patient and public representatives. However, we have identified six areas where further work is required to develop a productive ecosystem that that meets the changing needs of health data science to deliver public benefit whilst protecting individual privacy.
-
Consistent and proportionate accreditation of safe people.
-
Consistent accreditation of safe settings, making use of existing standards and with a focus on the use of public cloud computing.
-
Involvement of public and patient representatives in the data access management decision making process, with transparency of use, outcomes, and impact.
-
Improved lay explanations of the design and functioning of TREs.
-
Enhancing the researcher experience whilst minimising risks to privacy.
-
Addressing the technical, governance and process challenges of federating TREs.
Status of the document¶
This Green Paper has been updated to reflect the comments from a consultation run between 30 Apr and 26 May 2020. We are very grateful for the twenty-four responses received which covered a broad range of stakeholder views including patient and public representatives, NHS and other data providers, academia, system-level stakeholders, TRE service providers and consultancies. Whilst it has not been possible to incorporate all of the comments received, the input will also be used in the development of the implementation plan.
We continue to welcome comments and contributions to help shape the development and subsequent policy papers on the different areas outlined above.
A summary of changes to the previous draft can be found at Appendix B.
Overview¶
Purpose¶
Health data used for research and innovation comes from a variety of sources, but most relates to peoples' interaction with the health and care system in some way -- for example as an NHS patient, a participant in a clinical trial, being involved in a genomics initiative or as a blood donor. Therefore, achieving the confidence and trust of patients and the public in the use of this data is central to achieving our vision.
This paper provides the case for a shift to providing access to data via Trusted (or Trustworthy12) Research Environments (TREs) which protect - by design - the privacy of individuals whose health data they hold, while facilitating large scale data analysis using High Performance Computing that increases understanding of disease and improvements in health and care. It sets out the requirements for TREs based on the Five Safes13 model, with some extensions to reflect the latest technological developments and specific requirements of health data. It sets out a potential direction of travel for the transparent, independent accreditation of TREs that will make up the infrastructure of the UK Health Data Research Alliance, as well as how federation across this 'national grid' of TREs could occur. It concludes by identifying six work packages required for implementation.
Background¶
The UK Health Data Research Alliance is an alliance of leading health, care and research organisations united to establish best practice to enable the ethical use of UK health data for research and innovation at scale14. A central challenge in using health data is how to facilitate research while protecting privacy and so engendering public trust.
The Office of National Statistics (ONS) which facilitates research access to similarly sensitive administrative data described in 201715 its role as to "find a way to maximise the use of the detailed data that ONS holds, while keeping them secure at all times; to let government, academics, businesses and others use these data, while being able to assure you [the public] that you will never be identified, your private details will never become public and that the information you have given us will only ever be used in ways that clearly serve the public good". Their approach is summarised as "Five Safes": Safe people; Safe projects; Safe settings; Safe outputs; Safe data. These "Five Safes" should be considered as adjustable controls rather than binary settings. Risk is addressed by complementary adjustments on the implementation of each "Safe" to provide an appropriate context for research to occur which maintains an optimal balance between research benefit and overall risk management.
The term "Data Safe Havens" has been used to describe systems for providing researchers with access to data while managing risk of unauthorised re-identification of individuals from de-identified data, however implementations vary considerably16. When evaluated against the "Five Safes" defined by ONS, common features of Data Safe Havens are processes of evaluation of research proposals before granting access (Safe projects) and robust processes to de-identify and anonymise data being accessed to reduce the risk of re-identification (Safe data). However, in many cases the model of access is one of "Data Release", i.e. where processed datasets are distributed to researchers, rather than requiring them to carry out their analysis within a controlled environment operated by the Data Safe Haven (Safe setting). Once data is distributed to researchers, controls on who accesses the data (Safe people) and on what is publicly released as results (Safe outputs) and even exactly what research the data is used for (Safe projects) are out of direct control of the Data Custodians.
Historically organisations such as ONS that have operated safe settings have offered only a limited set of statistical analysis tools to researchers accessing data in their environment. These have been largely adequate given the structure and size of administrative and social science datasets being analysed. By contrast, health datasets, consisting of electronic health records, assets/images, and omics data types, are typically richer and larger. Analysis is more likely to require complex custom analysis algorithms which correspondingly require more substantial compute resources.
Until recently it has been a challenge to provide a safe setting able to support analysis at such a scale and with such diverse tooling. There has also been a history, dating back to the open sharing of the first human genome, of data distribution for such biological datasets to researchers to allow them to carry out analysis on their own computer systems. As a result, with the rise of the generation of omics data from consented individuals, this data distribution model has become codified as the process for managed data sharing, exemplified by the archives database of Genotypes and Phenotypes (dbGaP) and European Genome-phenome Archive (EGA).
Critical to the success of the proposed TRE-based approach will be achieving the optimal balance between confidence of data controllers through increased security, benefits to the researcher through improved access to larger datasets, and transparency for public and patients as to who is accessing the data and for what purposes.
A reduction in researcher productivity has been highlighted as a risk through the initial consultation. It will therefore be essential for the UK Health Data Research Alliance TRE workstream to continue to engage the community to develop best practice for the full range of researcher requirements and experience. It will also be important to communicate the other benefits that will accompany a move away from the data release model such as improved data access request turnaround times and approaches to enable greater potential for hypothesis-generating or agnostic analysis.
The case for TREs providing access to health data through safe settings¶
Recent events and developments have made the provision of safe settings for health data analysis both a desirable and technically practical alternative to data distribution.
Firstly, previous initiatives that have not effectively engaged or consulted on the rationale for data access have resulted in a lack of trust in the phrase 'data sharing'. It has become associated with the risk of jigsaw reidentification through the distribution of data to unknown third parties for unknowable types of analysis and potential unknown further distribution and linkage. All approaches to the de-identification of datasets that contain individual patient level data are limited and require controls enabled by TREs.
Secondly, the adoption of General Data Protection Regulation (GDPR) has made researchers and their organisations subject to serious financial consequences of failing to adequately protect personal health data distributed to them and hosted on computer systems they are responsible for. For many organisations, in particular universities and NHS trusts, it has becoming increasingly challenging to operate computing environments with the required level of security. This is particularly the case when large scale high-performance computing (HPC) is required to support the research community. Similarly, data custodians distributing data have become more risk averse because of potential shared responsibility with receiving organisations for any breach under GDPR.
Thirdly, for large datasets such as assets/images and genomes, data distribution is both inefficient and costly. It results in funders directly or indirectly supporting the costs of storage of multiple copies of large datasets and the associated network costs for multiple large data transfers, when each copy may only be used for a limited period.
Fourthly, over the last 5-10 years the evolution of computer systems have made it practical for researchers to bring complex analysis pipelines to data held on centralised systems17 and for these systems to be able to support both cost efficient and dynamic scalability of compute18 for analysis and integral data security19.
Finally, there are examples of operational systems that provide a safe setting as the only mode of access and that are being used at scale for these classes of health data. The SAIL DataBank has operated for 12 years with a 'no data leaves' or 'reading library' approach to data access. Using Swansea University's UK Secure e-Research Platform (UKSeRP) to deliver remote access to population-scaled linked data resources from over 400 partner organisations, UKSeRP's fully featured high powered analytical environment has supported hundreds of projects conducted by academics from across the UK over that time20. Similarly, Scotland has implemented a system of federated safe havens with secure analytics platforms, which are safe settings, as described in their Safe Havens Charter21. The more recent Genomics England Research Environment (GERE) is also a safe setting and, with >20Pb of genome data from the 100,000 genomes project, operates at a much greater scale. It has >2,000 researchers onboarded to carry out analysis with a range of tools and a full HPC environment (see figure 1).
GERE has the advantages of being set up 1) with explicit research consent from each patient participant; 2) with substantial public and patient engagement and oversight and 3) with completely unequivocal published22 and public statements that individual level data will not be distributed but will remain within the research environment. As such it has achieved a high level of trust despite dealing with individual genomes, a new and sensitive class of personal health data23.
UKSeRP has been available as a private cloud offering to third parties wishing to take advantage of UKSeRP's "ready to go" platform to secure and provide access to their own data, under their own governance 24 25. As of the 1^st^ January 2020 there were 25 UK-based organisations with UKSeRP tenancies in the UK, including Dementias Platform UK, Avon Longitudinal Study of Parents and Children (ALSPAC), and UK-CRIS (Clinical Record Interactive Search), with an increasing number of installations internationally.
It is apparent from patient and public engagement work that there is much greater comfort with data being accessed through a safe setting than data distribution.26 Where research access is via a safe setting with appropriate patient /public oversight of research activities an opt-out consent model may also be considered sufficient even where data is sensitive, such as the founder CRIS system providing a research environment with Natural Language Processing (NLP) tools on a de-identified copy of the EHR records of the South London and Maudsley NHS Foundation Trust (SLaM)27. Even during past media-fuelled public concern about data sharing, the SAIL Databank with its robust proven and robust approach to data curation and access remained uncriticised28.
Most recently, the OneLondon Citizens' Summit Public deliberation in the use of health and care data identified the following factors that reassured participants in relation to data access29:
-
Research organisations accessing data within a controlled and secure environment, such as a hospital or research hub, and the data not leaving this environment;
-
Access being supervised by appropriate NHS staff or conducted by NHS analysts on behalf of the research organisation;
-
Contractual arrangements in place that underpin the data access with consequences for those who break the rules around access (e.g. sharing data outside of the research environment);
-
Data not sent or shared outside of the research environment (but could be accessed remotely).
Requirements for a Trusted Research Environment¶
The UK Health Data Research Alliance's Principles for Participation30 include use a proportionate approach to the governance of data access based on the five "safes". TREs provide a "safe setting" approach. For the Alliance to maximise its potential, common agreed specifications will simplify processes for researchers, lowering barriers to access to multiple TREs. Over time, a common TRE specification(s) combined with the adoption of common health data standards will facilitate federated analysis across multiple TREs.
Ensuring public trust is maintained across multiple TREs implemented in different ways and operated by different organisations will require both the adoption of a common TRE specification and independent accreditation and auditing (see accreditation below).
Most Alliance members manage health service data for which access through the safe setting model is most appropriate. A subset of members manage access to research cohorts where rules of data access may be significantly different. This is due to participants in research cohorts being volunteers who have consented to data access rules that were ethically agreed at the outset. For example, the consent of one of the largest UK research cohorts, UK Biobank 31allows data distribution to approved researchers ("safe people") who have an approved research plan ("safe projects"). For these members setting up a TRE instance based on the safe setting model need only be one method of data access. However, to maintain the integrity of the ecosystem, such a TRE instance would need to be completely isolated from systems that provide data release.
Safe people¶
Individuals allowed access to TREs should be researchers32 able to demonstrate appropriate credentials and be undertaking approved (safe) projects. They are likely to be paid by research organisations that are prepared to take responsibility for their actions and vouch for each individual. However, the approach must not inadvertently constrain access for researchers from non-standard backgrounds, such as those bringing distinctive data science capabilities from other sectors such as social sciences, finance or from start-ups. Researchers would be required to sign legally binding terms of use including:
-
not trying to re-identify individuals33
-
immediately reporting any security weakness found when using the system and not attempting to exploit it
-
not sharing their login credentials with any other individual
-
informing the TRE if they are changing institutions before they have done so
They would also be required to carry out information governance training and, potentially, training specific to the health domain, TRE and/or datasets, refreshed periodically.
Accredited TREs will require systems to track individuals and organisations, the status of their on-boarding progress. With a common definition for safe people across the Alliance, it would be possible to setup something like the "ONS Approved Researcher Scheme" or "Accredited researcher under the Digital Economy Act 2017"34 , where researchers only have to be approved once to access multiple TREs, subject to separate project approvals. This is also the approach that is being developed through the Global Alliance for Genomics and Health (GA4GH) with their passport and visa standards35.
The Health Data Research Innovation Gateway (the 'Gateway')36, working in concert with the ONS Research Accreditation Service37 has the potential to provide the technical implementation to reduce burden on researchers, and their parent organisations, wanting to access multiple datasets across different Alliance TREs. This would also simplify implementation for each TRE and would be achieved either through the Alliance managing identity of researchers itself or relying on third party national or international research directories that have been proposed should they implement appropriate review and management processes.
Summary of Consultation responses to question: Safe people: how do we accredit researchers in a way that can support academia, NHS and industry to achieve the vision that every health and care interaction and research endeavour will be enhanced by access to large scale data and advanced analytics?
- General support for the need to have an open standard that may need to be adjusted for specific datasets and specific TREs.
- Should support a tiered approach to accreditation aligned to different levels of data sensitivity
- Prefer licensing rather than one-off accreditation with a central body providing licencing.
- Use existing accreditation schemes (e.g., aggregation of many of the common "tokens of trust")
- Risks to mitigate included:
- Accreditation of individuals may be cost prohibitive, could fall under a broader research unit or organisational accreditation.
- Accreditations and approaches become nothing more than box ticking exercises.
Safe projects¶
Despite the privacy protections offered by TREs, it remains essential to ensure that the use of data is appropriate and has the potential for public benefit. The TRE must have the functionality to enable this use to be audited to ensure compliance. Alliance members already have systems in place to review proposed research projects, and the functionality of the Health Data Research Innovation Gateway is being developed to support the harmonisation of these systems as far as possible.
Involving representatives of patients and public is a key element, alongside transparency of decision making and data use, as highlighted in the Foundations of Fairness joint report from Understanding Patient Data and the Ada Lovelace Institute38 and subsequent learning data governance model proposals39. This includes recommendations to address the issue of frequency of feedback and transparency around the research that is being carried out, that patients, public and cohort research participants frequently raise in focus groups. To improve transparency, it is proposed that TREs require lay summaries to be provided as part of the project approval process with these being made public on approval. TREs should also implement systems to allow access by researchers of data held by the TREs to be linked back to projects so that research outputs can be made transparent and reported back to participants as standard.
Further consideration of safe projects will be considered as part of the Data Access management module development of the Innovation Gateway.
Safe setting¶
As detailed above, there are multiple existing platforms providing research access to health data through the implementation of a safe setting. While one side of operating a safe setting is the need to ensure public and data controller trust through security and transparency, the other side is the need to ensure it is engineered to be as easy to use for research as possible.
At minimum a safe setting needs to implement:
-
A system to hold data securely such that individual level data cannot be exported. For transparency the security design and implementations should be independently audited with reports reviewed by a patient/public oversight group and made public.
-
Systems to allow secure remote access by accredited researchers to carry out analysis with the ability to keep track of researcher activity (to ensure compliance with "safe projects") and that ensures accounts cannot be shared (to ensure compliance with "safe people").
-
A research environment containing a set of tools to allow data to be analysed, with a barrier between the safe setting environment and the outside world to control data and software import or export, referred to below as the 'air lock'.
-
Processes and systems for export of summary data (Safe output, see below) and to support data or software import. It will be necessary to implement systems able to scan data files, such as for viruses hidden within software packages and for identifiable data that should not be imported into a TRE.
The types of analysis that researchers wish to carry out may go beyond statistical packages provided as standard by TREs (e.g. R-Studio, Stata). It must therefore be possible for researchers to bring their algorithms into the safe setting. The 'Air lock' capability ensures that imported tools are scanned to check that they will not compromise the security and integrity of the TRE and, in particular, do not facilitate the export of record level data.
Similarly, researchers may wish to analyse data in the safe setting along with data held outside. The safe setting must provide mechanisms to support importing and linkage of data. As with tools, this will require 'air lock' capability to allow for the secure importing of researcher supplied data. This 'air lock' may require tools to ensure that this data does not compromise the environment nor enhance the risk of re-identification beyond that assessed at the time the researcher's request for access was approved.
Data linkage will also require a mechanism to undertake the linkage in such a way that privacy is maintained or enhanced, such as using the services of a trusted third party.
In practice, such controls mean that researchers cannot operate inside the safe setting as they do on their own computer systems. While they can access the safe setting remotely via a Virtual Desktop Interface (VDI) to carry out their research, they will not be able to connect from the safe setting to the outside except via the 'air lock'. This means they will not be able to access external websites, unless these are whitelisted with appropriate security to make them read only to prevent data export. It also means they cannot easily connect directly to software repositories such as GitHub.
To facilitate the development and configuration of software prior to import into the safe setting, it is likely to be beneficial for TREs to provide researchers with a separate test environment that behaves like the safe setting, but is accessible from the internet. Such a test environment would need to have the data frameworks identical to those within the safe setting so processing of data formats and communication to APIs could be tested, but the data itself would need be synthetic40. Since software frequently assumes the existence of the internet and contains embedded external file requests etc. it is also sensible for this test environment to have a 'no-internet' mode so it can be checked that the software still runs in the absence of internet connectivity.
TRE safe settings will be multi-user environments, in most cases with the ability to run algorithms on high performance computing systems, either as on-premise native HPC, private cloud or public cloud (see safe computing below). In all cases systems to manage the competing demands of many researchers are essential, meaning imported software must be able to work with such systems. Similarly, regarding actual software import, software could be packaged in different ways including containers (such as Docker) to full virtual machines. Across the Alliance it may be appropriate for different TREs to consolidate on supporting one or a limited number of workflow management and software packaging solutions to reduce the complexity for researchers wanting to run their software in different TREs.
Safe computing -- an extension of Safe setting¶
Since the ONS definition was originally developed, a new issue has become important that is not explicitly covered by the "Five Safes" but needs to be addressed to build public trust. This is the outsourcing of provision of computing infrastructure for all or part of a safe setting to third parties through partnerships with commercial organisations or use of public cloud computing providers.
Previously, safe settings have been almost exclusively provisioned through "on-premise" computer hardware where physical security of equipment, network security, software maintenance etc. is the responsibility of the data custodian or TRE operator. Such systems can be configured as "private cloud" to support the use of software distributed as virtual machines and containers. However, use of third-party computing resources, such as public cloud, offers many potential advantages for TRE providers and is likely to be the default from now onwards. This provides dynamic scalability of compute to enable short periods of intensive computation such as for AI training. Outsourcing layers of the hardware and software stack which have become commodities to cloud providers brings other potential benefits due to their greater capacity to engineer scalable platforms and implement robust security.
In order to build public trust, use of private sector computing infrastructure to provide a safe setting must be done in such a way that none of the hardware and software layers outsourced make it possible for the third-party provider to access individual health data. This needs to be enabled through security design and engineering as well as contractual arrangements with the third-party provider to minimise the risk of a data security breach. It is accepted by some cloud providers that a security design that ensures they have no data access is a critical requirement to many organisations. Technical papers have been published about how to engineer this level of security where cloud provider administrators have no ability to access any customer data41^,^42.
The security engineering and design required to make this possible is complex and involves encryption on rest of all health data and encryption key management infrastructure configuration such that only the data custodian controls the keys. It is proposed that TREs using public cloud should be engineered in this way and would be regarded as operating a "safe setting" that implements "safe computing".
Explaining this complex engineering and design in ways that data custodians, researchers and members of the public can understand and engenders trust represents a challenge that needs further consideration.
Summary of Consultation responses to question: Safe setting: How do we provide assurance that Trusted Research Environments that make use of public cloud storage and computing are safe settings and that data cannot be accessed by the hosting technology companies?
- Manage public cloud providers by contract, reputation and physical controls (keys).
- Require assurance at technical level for security standards; at systems level for robust ISMS provision; at governance level in terms of data sharing agreements.
- Require specific information controls including encryption at rest with key management segregation so that the hosting company can be shown to be unable to access data.
- Require all processes that generate keys derived from customer-managed keys be audited, with alerting in place for unexpected access.
- Apply permissions technology that couples data use(s) to data access.
- Require transparency around service level agreement and support agreement, including the conditions under which its employees would need to access encrypted volumes.
- Examples provided included: Amazon AWS, Microsoft Azure and Google GCP all offer FIPS 140-2 compliant key management services backed by hardware security modules (HSMs); NHS Digital "Health and Social Care Cloud Security - Good Practice Guide“43; Intel's SGX secure compute extensions; IGToolkit/DSPT, ISO27001, Cyber Essentials, Data Protection Impact Assessment (DPIA); ONS / UKSA Accredited Processor.
Summary of Consultation responses to question: Safe setting: How is this [use of public cloud] best explained to data custodians, researchers and the public and not just security experts?
- Very strong support for shift to public cloud: recommendation to use NHS Digital's language to signal a more positive attitude towards its use.
- Cloud host should implement a layer of transparency to help assure public that public cloud solutions are safe and well managed, making available its agreements, including the conditions under which its employees would need to access encrypted volumes; publishing accreditation met, results of audits and improvement plans/corrective action plans,.
- Language used must be consistent and clear without assuming specialist or prior knowledge. Visual representations can be a powerful and effective. Working with patients would help achieve this.
- Clinicians are crucial stakeholders who should be included in efforts to consult with patients and the public on the model being developed
- Communicate clearly the benefits of public cloud platforms to research in terms of flexibility, functionality, near-global accessibility and cost.
- Rather than explain, demonstrate physical and logical controls with total transparency by walking stakeholders through the process of data flow and access. Visits to UK facilities showing the physical data flow from data ingestion, data landing and data processing.
Safe data¶
As well as all the controls outlined above, TREs should ensure that the data accessible to researchers within the safe setting is proportionate to the approved project requirement, in line with GDPR requirements. Processes to import data into the safe setting include de-identification to minimise the risk of accidental re-identification of individuals. Best-of-breed de-identification and encryption, will replace the current 'data release' approach to anonymisation that either increases privacy risks or negatively impacts research analysis as a result of the anonymisation required to manage the risk of 'jigsaw re-identification'44 outside of a TRE. These limitations are central to the need to move towards a TRE based model for future research and innovation.
However, with the additional controls supported by the other "Safes" current conservative approaches to linkage and data minimisation can be adjusted therefore opening up new research opportunities and especially facilitating a more effective approach to hypothesis-generating research that has the potential for public benefit. Environments such as GERE which implements a safe setting with strong implementation of the other safes, allow researchers to analyse across the entire dataset, facilitating broad investigations of genome/phenome relationships, multimorbidity effects etc.
Safe outputs¶
As outlined in Safe setting, TREs must implement a barrier (or "air lock") between the safe setting environment and the outside world to prevent unauthorised data export (or import). TREs must implement processes and systems to allow approved data to cross this barrier. Systems require functionality to track requests and decisions, supporting cycles of rejection and revision.
Current approaches to review requests to export summary data are based on manual review with typically final release being governed by an oversight committee. This is a potential bottleneck and could be one of the factors that will negatively impact the experience of a researcher moving from a locally hosted analysis environment to a TRE. Work is therefore required to explore approaches to automation or partial automation where the risks of disclosure can be adequately controlled. This will also be beneficial for the data custodians to make management more scalable and sustainable. There are opportunities to establish a network of TRE airlock managers to share expertise and develop consistent approaches to definition of safe summary outputs.
Summary of Consultation responses to question: Safe outputs: How do we achieve a scalable and trustworthy approach?
- Focus on simplicity and automation to enable scalability – the more manual the more potential for error, inconsistency, and delay.
- Adopt holistic approach to output management: training of researchers and categorisation of outputs which are safe or require further manual assessment.
- Aim at reaching point of trusting researchers and your system for assuring they are trustworthy.
- Develop ability to approve reusable output pipelines as safe rather than individual instances of output data – output reviewers need to be appropriately trained.
- Consider tiered risk assessment – automated processes to scan outputs and classify asset and associated risk. Low risk could be under control of Principal Investigator.
- Helpful if team in charge of data feed also responsible for checks to scan outputs.
- Can be achieved with the right type of output policy and careful management of researchers’ needs.
- Safe egress of individual level data only to another TRE with equivalent level of security.
- Learn from ONS, UK Data Service, HMRC DataLab and GERC that already implement safe outputs for their hundreds of researchers
- Implement controls for reputational risks as well as statistical disclosure.
- Consider different purposes: ONS identifies 3 distinct outputs:
- Pre-publication clearance
- Publication clearance
- Code file clearance
Safe return - Extending the Trusted Research Environment definition¶
While health data held within TREs is de-identified for most research purposes to guard against accidental re-identification of individuals by researchers (safe data), there are differences as to whether it is consented and/or technically possible to send individual analysis results back to the clinical setting that originated the data and where identities are known. This will be for individual clinical care purposes and invitations to participate in trials and other research projects.
For example, in the Genomics England case (see figure 1), there is ethical approval and patient consent to pass analysis results for an individual generated in the research environment safe setting back to the clinical setting for re-identification, evaluation and return for clinical care. These 'outputs' supplement the results already generated by clinical analysis pipelines in the clinical setting. Given that the clinical analysis pipelines only produce diagnostic results in 20-25% of cases, there is considerable clinical value in additional individual diagnoses being proposed for undiagnosed patients from the research side. Making this possible requires completely robust and certified data paths for individuals to ensure that a result obtained in a research environment is always perfectly mapped back to that individual's clinical record.
On the other hand, in the case of research cohorts such as UK Biobank, which does contain clinical health data for each individual, there is no consent for return of results to individuals so a TRE based on UK Biobank data would not allow this.
For TREs where return of results is possible, there can be multiple benefits. It may be only a part of the research activity carried out within a TRE, but supporting this option has the potential of increasing the convergence of research and clinical care, bringing researchers and clinicians closer together. It may also provide an additional incentive to clinicians to ensure the clinical data they record is as complete as possible if research use could result in additional clinical feedback.
Implementing such a return path in a way that ensures no reports are returned to the wrong individual requires a trusted linkage service to manage the keys and for the receiving body to manage consent. There was a split view on whether to consider this a separate safe or whether it should be treated as extension of 'safe output'. There was, however, broad agreement that a tiered approach to TREs on a number of dimensions should be considered and that this was only one part of functionality.
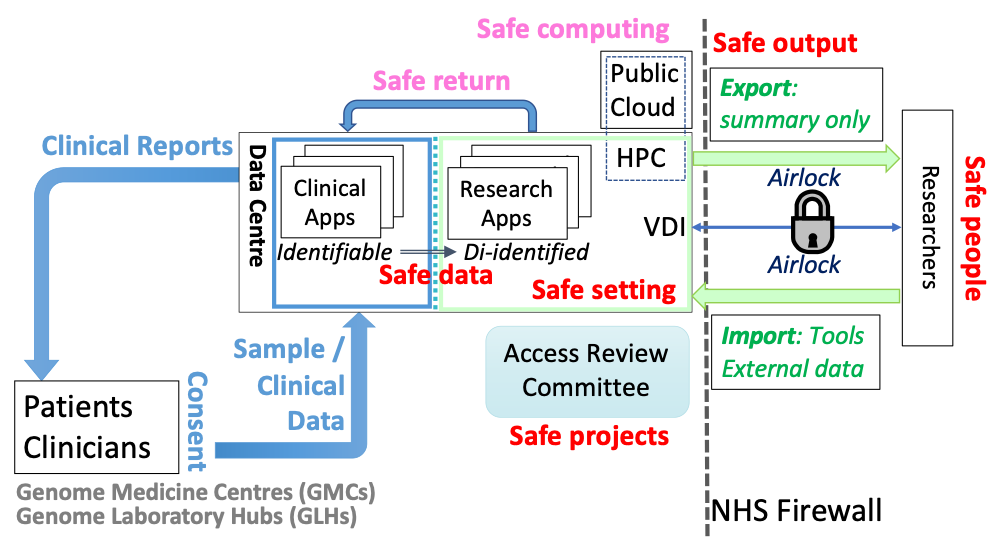
{#Fig1}Figure 1: Schematic example of a TRE+ environment. The Genomics England TRE operates as a safe setting [green] that approved researchers (Safe people) can only access via a virtual desktop interface (VDI). Only de-identified data is accessible by researchers (Safe data). Research is overseen by an Access Review Committee (Safe projects). Only summary data can be exported from the TRE through an Airlock and only after manual review (Safe output). Analysis of genome data requires High Performance Compute (HPC) resources, however scaling HPC to meet the needs of large numbers of researchers is challenging. One solution would be to also use public cloud resources; however, this would need to meet the security requirements of Safe computing. In the Genomics England environment, the data accessed by researchers is a de-identified version of real-world data being analysed to produce reports for clinical care [blue]. This makes it possible to pass back research results that may be relevant for individual clinical care (Safe return).
Summary of Consultation responses to question: Do you think this particular output [safe return] should be considered a separate ‘safe’ and that this functionality would represent a TRE + model
- The Safe Return discussion needs to consider the patient safety implications and the associated use of clinical risk management standards for both (i) manufacture and (ii) application and deployment of health IT systems respectively (DCB0129 and DCB0160) to mitigate any safety concerns
- “Safe returns” is a fundamentally problematic notion that should be removed, and intent achieved through existing and legitimate relationships
- We have been required by our customers to provide re-identification services as part of TRE implementations as standard.
- The concept of 'Safe Return' is excellent, and much needed….[but] we think actually that it could constitute an additional 'safe output.
- There should always be a secure way of communicating relevant research results to the data custodians who submitted the data to the TRE, if the data is reversibly psuedonymised. It is their decision on whether to act or not.
Researcher requirements for a TRE environment and for data federation¶
Since the objective of setting up TREs is to enable research on health data at scale, it is important that the implementation of the "Safes" as previously described do not unnecessarily restrict or slow down research activity. As previously described, there are many advantages, such as in terms of trust, security and cost, of providing a single controlled point of access to data through a TRE, however there are also risks of lower research productivity if researcher concerns about difficulties of carrying out analysis within a TRE are not addressed. For example, one legitimate researcher concern is that TREs have sufficient support staff and/or sufficiently automated systems to ensure rapid import of data; installation of software and export of summary results to minimise research delays. Other concerns may be less specific to TREs, such as the current lack of expertise in deploying software in cloud environment. Training or support for researchers is clearly needed to address this, but here TREs are not unique, just at the vanguard of adoption of public cloud that will ultimately affect all researchers.
For this reason, engagement with researchers is critical. One of the six proposed follow-up work packages is about enhancing research experience while minimising risks to privacy and the consultation included a question specifically about functionality required:
Summary of Consultation responses to question: User centred functionality: What is the minimum set of analytical tools and functionality that a TRE must make available for researchers?
- Requirements for tools will be domain specific. This is where TREs can differentiate themselves
-
Researcher engagement should not be limited to giving specifications. Researchers should be embedded in the entire TRE environment development process.
-
Key requirement for a TRE that seeks to provide a productive, highly usable analysis environment is a smooth, fast process for reviewing, approving and provisioning new software and tooling within it.
-
Open source scientific software is often not mature enough to run in cloud or hosting platforms using standard security models (e.g. OAuth2) and data management (shared hosted disks, blob stores). HDR UK could support funding software developers to make their tools more secure and robust for use in TREs.
-
TREs will benefit from being able to access large virtual machines, or virtual machines equipped with graphical processing units.
-
A benefit of TREs is ability to dial up computing power as needed, however mechanisms will be required to manage spending.
Example tool requirements:
-
Graphics tool, statistical tools (e.g., R, RStudio, Stata), Programming language (e.g. Python, Perl)
-
Data Visualisation tool (e.g., Tableau), Data Integration tool
-
Version control client for collaborative working, chat functionality.
-
Browser, file compression, text editor, word processor, spreadsheet, presentation, PDF Reader
-
SQL database (e.g., Postgres)
-
For genetics: Workflow engines (e.g., nextflow, WDL), high-performance general-purpose compute with APIs and CLIs
-
Jupyter Notebooks for reproducible code
-
Software services for data munging, stratification e.g., cohort browsers
-
Access to package repositories (e.g., CRAN, PyPI, BioConductor, Conda) and fast process for approval and installation.
A second related researcher concern is about being able to carry out analysis across multiple datasets when the TRE model does not allow distribution of individual level data. When a second dataset can be distributed (such as a research cohort) it may be possible to import that dataset into the TRE to carry out analysis across the two datasets there. However, if the same restrictions apply to both datasets, then some sort of federated analysis between multiple TREs will be required. In the case of researchers wanting to carry out analysis across health system data from multiple countries it is likely that that most countries will not allow individual level data to cross national boundaries (unless part of a research cohort) making federation the likely future norm for this type of research.
The simplest form of federated analysis involves running analysis algorithms separately on the data held within each TRE and then exporting resultant summary outputs (safe outputs) and combining them. Such an approach is already used in cases where individual level data cannot be directly combined, such as meta-analysis of genome wide association studies. Similarly, there are approaches where TRE environments provide programmatic interfaces that implement the summarisation rules of safe outputs, enabling researchers to carry out limited types of federated queries remotely across multiple TREs without having to install and run software on each. Implemented examples include the beacons network45 developed by the Global Alliance for Genomics and Health46.
However, there are classes of experiment that may benefit from a tighter form of data federation or the aggregation of datasets to allow linking within a single TRE or virtual TRE environment. One way of supporting this is to build upon an accreditation process for TREs (see below) to establish a common model of trust across a federation of TREs with common researcher identity. Not all TREs may implement the requirements in this paper to the same level and therefore a potential asymmetric hierarchical model of data flow may be required, ensuring that aggregated data is accessed in the safest environment across the federation. This could build on experience from the security and defence sector47 and the implementation of the Digital Economy Act which categorises two levels of processing covering preparation (including linkage) and provision, and provision only.48
A move towards federated analytics and distributed Machine Learning will need to be underpinned by such a federation of TREs. However, it is likely to be some time before this approach can be fully realised and in the interim the default approach will need to be through data federation utilising techniques such as from the HDR UK Sprint Project -- "Graph-Based Data Federation for Healthcare Data Science".49 This would then support the controlled transient aggregation and linkage of data across TREs to support data federation. This will require auditability of data, secure transfer mechanism and controlled destruction of the transient data in line with the requirements of ISO 27001.
There are many technical, governance and process issues connected with the development of systems to support federation between TREs across the HDR Alliance and more broadly. For this reason, the final proposed follow-up work package concerns this challenge.
Accreditation of TREs¶
This paper has outlined a proposed approach for TREs. However, for such an approach to be implemented, it will need a widely accepted accreditation process that meets the requirements of data custodians, regulatory bodies and patients and public representatives. At the same time, this will need to be achieved without an excessive additional burden on the TRE who may already be undertaking other accreditation standards.
Consensus is needed on who will provide accreditation, what is within scope (characteristics, processes, personnel, etc.) and how this will build on existing frameworks such as ISO 2700150, the NHS Digital Data Security and Protection Toolkit (DSPT)51, and the UK Statistics Authority accreditation process that supports implementation of the Digital Economy Act (DEA)52.
Much work remains to be done in this area and there are challenges, such as the highly technical oversight of security design and implementation required to support safe computing on public cloud.
Summary of Consultation responses to question: What approaches should be considered to assess that TREs meet the characteristics required?
-
Independent, 3rd party audit to a known, recognised standard, along with NHS DSP Toolkit audit.
-
Some international standard with external audit - accreditation process must not inhibit international collaboration.
-
Rely on existing certification schemes.
-
Scalability can be advanced by a national use case working group (UCWG) which documents the data use case, the IG and technical spec once (do once, well and share principle)
-
An application to demonstrate and provide evidence for how the TRE meets the criteria with review and on-going audit to verify the information provided and explore areas of concern. The review and audits would require a team with multi-disciplinary skills.
-
Specific set of recommendations interpreting the current standards in health data context (e.g., approach taken in NHS Digital "Health and Social Care Cloud Security - Good Practice Guide “)
-
Self-certification with requirement to upload evidence.
Examples
-
Minimum: cyber essentials plus; ISO27001; NHS DSP Toolkit compliant
-
ISO27001, HITRUST
-
Safe-Box - Safe-Haven
-
National Cyber Security Centre
-
ONS Digital Economy Act Accreditation
-
Maturity model approach (like HIMSS or Energy Rating)
-
Alan Turing Institute sensitivity tiers.53
-
5 Safes could be used as the accrediting framework
-
Combination of ISO 27001 (Information Security) and ISO 9001 (Quality) tailored with inclusion of principles taken from both DCB0129 and DCB0160 (Clinical Safety Standards).
Summary of Consultation responses to question: Who should be the accrediting authority? How should this be funded?
- Health Data Research UK or affiliated body (e.g., UK Health Data Research Alliance) x 5 mentions
- ONS / UK Statistics Authority x 4
- NHSx, NHS Digital x 2
- The Health Foundation
- ‘Existing independent body’
- Industry association e.g., Data Centre Alliance, DC Uptime Institute
- MHRA
- UKRI to establish a multi-sector, cross-disciplinary function.
- Work with ISO to define an international standard [and market for external auditors]
- National body plus local responsibilities.
Funding?
- Organisation offering the TRE (x 4 mentions)
- UKRI
- Proportionate membership fee for one or more of data providers, research funders and TRE providers.
- UK Health Data Research Alliance or HDR-UK if low financial cost (higher in-kind burden)
Communications, Engagement and Involvement¶
Public Trust¶
Central to the adoption of the 'Five Safes' approach is the need to earn, build and sustain public trust. There is valid public concern over the control of data that is made available for research through data release and on the limits of de-identification. Communications, engagement and involvement with the public must be central to the Alliance's proposed approach to move towards data access via TREs, implement accreditation and support integration with the Health Data Research Innovation Gateway. This disruptive change must respond to key public questions and concerns such that potential benefits from health data research are achieved whilst protecting privacy.
Communications¶
The workstream will need to ensure that there are clear and tailored communications for all stakeholders. This will need to address the questions and interests that are specific to the audience and the benefits that will accrue from a robust TRE model for research on health data.
Public and Patients -- Discussion needs to cover how the "Five Safes" augment de-identification, how this approach ensures that data remains under the control of the data custodians and not passed to private companies with the risk of use for unapproved purposes and that this will facilitate the UK as being the place for safe and secure health data research. Specifically, this will also need to cover the controls in place on data held in public cloud that ensures that the data cannot be accessed by the hosting organisation. Transparency regarding use and benefit of the data will remain paramount.
Data Custodians -- It needs to be assured that this will provide security for the data they manage and that it remains within their controls and meets GDPR and Common Law Duty of Confidence requirements. They will need to be able to support the approach to accreditation with confidence that this builds upon the data management requirements of ISO 27001 and DSPT. This enhanced level of control, compared to the data release model, should then encourage the data custodian to adjust their data access management processes to facilitate quicker access to richer data given the reduced risk of inappropriate disclosure or use.
Researchers and Innovators -- It will need to be assured that their user experience has been considered in the development of the requirements and that there is focus on a first-class research and innovation experience. The workstream will need to address the concerns from this community that a TRE approach will impact research efficiency. The communications will need to highlight other longer-term benefits such as more rapid access to data and improved opportunities for linking data that has until now been restricted due to the data custodians risk positions. A TRE model should also provide a more cost effective approach to high scale compute and storage as environments move to a hybrid cloud model and the cloud providers' commercial models are refined to address some of the current issues for example around the costs of data egress.54
TRE Service Providers -- The proposed approach offers service providers significant opportunities. But with these opportunities comes great responsibility to develop technical and governance systems that can protect privacy whilst providing a world class analytical experience.
Funders -- TREs offer funders of research a range of potential benefits. These include more efficient research through improved utilisation of storage and compute resources; a proportionate approach to data access requests based on all the five safes; and audit trails on provenance of research outputs and data manipulation. This supports both transparency and replicability. These features may also benefit regulators.
Summary of Consultation responses to question: What questions might we want to ask of patients and the public to enable an approach that builds public trust and helps develop a clear and engaging narrative about the benefits of health data research at scale and privacy protection afforded by TREs and the ‘five safes’?
- By being open and honest and facilitating genuine dialogue and not talking AT people
- By saying what you do and doing what you say you do, verifiably
- By being quick to tell the ICO / stakeholders when you make a mistake
- The use of a TRE is a small part of the business of building public trust - that research has a public benefit is a bigger issue.
- Honesty requires that you also explain the risks of TREs, especially that they have the potential for decreasing research productivity while not achieving substantive increases in data security.
- Explaining benefits and addressing concerns in non-expert language and figures to be effective when needing to build confidence with non-expert stakeholders
- Systems can be developed to provide individuals with a view on how their data contribution has impacted specific research outcomes.
- Feedback to individuals through at home and mobile devices gives a feasible opt out route without massive investment in re-consenting
- Exposure, transparency, and wider public engagement of the research taking place in these environments.
- A go-to reference case as to the benefits of such environments that has wide publicity - that is what we all collectively must aim for.
- Important to ensure that all these partnerships [with industry providers of software provision, business analytics, computing infrastructure] are subject to rigorous public benefit assessments, with clarity over why third-party providers are being chosen. E.g., uniquely well-placed to provide technical expertise, scalable compute or some other necessary and specific resource.
- Business model of any commercial third party involved in setting up, managing or using this infrastructure will need to be clear. Frequently asked question is “What’s in it for them?” As prominent tech companies profit from brokering and selling data: this should not be the commercial model for TRE partners if they are to be trusted with health data and this should be made explicit.
Example activities
- Opinion formers in society that might include broadcast and print journalists and vloggers
- Lay summaries and patient public engagement panels in the health research space a great practice
- Participant led forums in e.g. Innovative Medicines Initiative (IMI) programme, European prevention of Alzheimer’s dementia (EPAD) and in the `#datasaveslives campaigns
- Diffusion of the benefits of data analysis on TREs through normal media channels e.g., targeted communication pieces posted on the NHS website or through media channels
- Targeted messaging at the point of consumption e.g., posters on the scientific results achieved thanks to the TREs in hospital wards and GP practices that are contributing data.
- Consulting Understanding Patient Data is a good place to begin
- COVID-19 response gives some good examples of how data processing in secure environments can work
- Via bodies such as the NIHR PPI teams and Comms, UKRI, etc. Research charities, especially Wellcome Trust; UPD; local charities.
- The Health Foundation has enlisted a PPIE (Public-Patient Involvement and Engagement) consultant to finalise holistic strategy.
- Connected Health Cities Manchester undertook separate engagement activities with different disease-specific patient communities in the local area
- Citizens juries - this is what Connecting Health Cities did for its TREs (each TRE was different but there were shared safeguards we checked ourselves against quarterly).
- We need a champion to do the PR, like Stephen Fry.
Next steps¶
Through the responses received during the consultation on the draft Green Paper and the workshops undertaken, we have identified the following six work packages that need to be put in place to support implementation of the proposed approach to TREs. In addition to working across members of the UK Health Data Research Alliance, including the health data research hubs, we also envisage working closely with the stakeholders outlined:
-
Consistent and proportionate accreditation of safe people. We intend to develop a shared set of criteria for identifying different tiers of safe people working closely with ONS and the Accredited Research Service and with MRC Regulatory Support Centre. This work package will also be developed alongside the HDR UK training strategy and seek alignment with the Researcher Passport and the GA4GH Passports and the Authorization and Authentication Infrastructure
-
Consistent accreditation of safe settings. This work package will be carried out in close coordination with ONS and the Digital Economy Act (DEA) Accredited Processor accreditation. UKRI and MRC planned investments in trusted research environments. This work package will not replicate existing accreditation approaches such as ISO 27001 or ONS accredited processor under the Digital Economy Act. It will focus on the implications for safe settings of the use of public cloud computing. As such, it will be important to work with cloud service providers as well as working across different sectors to draw on lessons from across sectors such as Finance, Criminal Justice, and Security.
-
Involvement of public and patient representatives in the data access management decision making process, with transparency of use, outcomes, and impact. This work package is already underway as part of the Innovation Gateway development of the data access management module. This work package is aiming to harmonise approaches to data access request across all different data custodians with a particular focus on those operating through TREs. This work includes consideration of public, patient and practitioner engagement and involvement in the decision-making associated with data access and the operating of TREs. Engagement with HDR UK Public Advisory Board, Understanding Patient Data and Use My Data and PPI groups of existing TRE operators will help guide this work.
-
Improved lay explanations of the design and functioning of TREs. Essential to effective communication and engagement is improved lay explanations and figures to explain the current situation and the proposed developments. Again, key stakeholders include HDR UK Public Advisory Board, Use My Data, Understand Patient Data, Association of Medical Research Charities and existing TRE operators. This work packages also picks up how information about TREs is represented on the Innovation Gateway. We will look to build on the work carried out by one London in their consultation on use of patient data.
-
Enhancing the researcher experience whilst minimising risks to privacy. User centred design must be adopted across the work packages. It is important that this includes academic, industry and NHS researchers and the service catalogue for TREs developed accordingly. Consideration will also be given to how researchers can provide objective feedback related to their experience operating within a TRE.
-
Addressing the technical, governance and process challenges of federating TREs. As outlined above, there are a range of challenges to address. This work package will need the engagement of existing TRE operators, data custodians, cloud service providers and researchers.
Appendix A: Consultation Questions¶
Appendix B: Summary of changes to document¶
References¶
-
https://understandingpatientdata.org.uk/how-do-people-feel-about-use-data ↩
-
For example: https://www.theguardian.com/commentisfree/2020/feb/16/our-personal-health-history-is-too-valuable-to-be-harvested-by-tech-giants, https://www.theguardian.com/technology/2020/feb/08/fears-over-sale-anonymous-nhs-patient-data ↩
-
https://www.hdruk.ac.uk/wp-content/uploads/2020/04/200416-COVID19-Research-Data-Final.pdf ↩
-
https://www.nhsresearchscotland.org.uk/research-in-scotland/data/safe-havens ↩
-
https://saildatabank.com/wp-content/uploads/UKSeRP_Brochure_v1.5.pdf ↩
-
https://www.genomicsengland.co.uk/about-genomics-england/research-environment/ ↩
-
https://www.ukdataservice.ac.uk/use-data/secure-lab/how-it-works.aspx ↩
-
https://digital.nhs.uk/coronavirus/coronavirus-data-services-updates/trusted-research-environment-service-for-england/ ↩
-
We have chosen to use the term 'trusted' but acknowledge there are other stakeholders who prefer the term 'trustworthy'. ↩
-
https://blog.ons.gov.uk/2017/01/27/the-five-safes-data-privacy-at-ons/ ↩
-
https://blog.ons.gov.uk/2017/01/27/the-five-safes-data-privacy-at-ons/ ↩
-
Burton, P. R. et al. Data Safe Havens in health research and healthcare. Bioinformatics 31, 3241--3248 (2015). https://europepmc.org/article/MED/26112289 ↩
-
Through Virtual machines or lightweight containers such as Docker. ↩
-
On premise High Performance Compute (HPC), private or public cloud services ↩
-
Data encryption at rest under data custodian control via Key Management Infrastructure. ↩
-
https://saildatabank.com/wp-content/uploads/SAIL_10_year_anniversary_brochure.pdf ↩
-
https://www.gov.scot/publications/charter-safe-havens-scotland-handling-unconsented-data-national-health-service-patient-records-support-research-statistics/pages/4/ ↩
-
Turnbull, C. et al. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ 361, k1687 (2018); Genomics England Protocol https://www.genomicsengland.co.uk/library-and-resources/ ↩
-
https://farrinstitute.org/wp-content/uploads/2018/03/UKSeRP_Case_Study.pdf ↩
-
https://saildatabank.com/wp-content/uploads/UKSeRP_Brochure_v1.5.pdf ↩
-
Great North Care Record 2018 Base: 824 North East representative ↩
-
e.g. CRIS system, allowing NLP over de-identified mental health records https://www.slam.nhs.uk/research/cris/ ↩
-
Lyons, Ford and Jones, Care.data: why are Scotland and Wales doing it differently? https://www.bmj.com/content/348/bmj.g1702/rr/687637 ↩
-
https://www.onelondon.online/wp-content/uploads/2020/07/Public-deliberation-in-the-use-of-health-and-care-data.pdf ↩
-
https://www.hdruk.ac.uk/wp-content/uploads/2020/03/200304-Principles-for-Participationv2pdf.pdf ↩
-
Researchers could be from academia, NHS and from industry. Any accreditation of research must cover all these communities. ↩
-
Data Protection Act 2018 Section 171 - Re-identification of de-identified personal data ↩
-
https://www.ons.gov.uk/aboutus/whatwedo/statistics/requestingstatistics/approvedresearcherscheme#becoming-an-approved-researcher-through-the-ons-approved-researcher-scheme ↩
-
https://www.ga4gh.org/news/ga4gh-passports-and-the-authorization-and-authentication-infrastructure/ ↩
-
https://healthdatagateway.org/ ↩
-
https://researchaccreditationservice.ons.gov.uk/ons/ONS_Homepage.ofml ↩
-
https://understandingpatientdata.org.uk/what-do-people-think-about-third-parties-using-nhs-data ↩
-
https://understandingpatientdata.org.uk/news/new-approach-decisions-about-data ↩
-
Synthetic data is artificially generated to replicate the statistical components of real-world data but doesn't contain any identifiable information. ↩
-
AWS: https://d1.awsstatic.com/whitepapers/using_aws_context_nhs_cloud_security_guidance.pdf; https://d1.awsstatic.com/whitepapers/Security/AWS_Security_Best_Practices.pdf; https://d0.awsstatic.com/whitepapers/AWS_Securing_Data_at_Rest_with_Encryption.pdf ↩
-
Google: https://services.google.com/fh/files/misc/handling_healthcare_data_uk.pdf, https://cloud.google.com/solutions/setting-up-a-hipaa-aligned-project ↩
-
https://digital.nhs.uk/data-and-information/looking-after-information/data-security-and-information-governance/nhs-and-social-care-data-off-shoring-and-the-use-of-public-cloud-services/ ↩
-
Jigsaw re-identification is the ability to identify people using two or more different pieces of information from two or more sources of information ↩
-
https://www.statisticsauthority.gov.uk/about-the-authority/better-useofdata-statistics-and-research/betterdataaccess-research/better-use-of-data/list-of-accredited-processors-under-the-research-strand-of-the-digital-economy-act/ ↩
-
https://www.hdruk.ac.uk/projects/graph-based-data-federation-for-healthcare-data-science/ ↩
-
https://www.iso.org/isoiec-27001-information-security.html ↩
-
https://www.statisticsauthority.gov.uk/about-the-authority/better-useofdata-statistics-and-research/betterdataaccess-research/better-access-to-data-for-research-information-for-processors/ ↩
-
Turing sensitivity tiers: pre-print paper https://arxiv.org/abs/1908.08737 poster https://doi.org/10.6084/m9.figshare.11815224 presentation: https://doi.org/10.6084/m9.figshare.11923644 ↩
-
https://www.gartner.com/en/documents/3939969/the-art-of-taming-data-egress-charges-in-hybrid-and-publ ↩


